Progress in 3-D Imaging Microscopy and Brain Mapping
"The ultimate goal is to better understand neurodegenerative, neurological, and psychiatric disorders such as Alzheimer's, Parkinson's, schizophrenia, and epilepsy and develop new treatments."
This was the mission of the national BRAIN Initiative launched in 2013, a vision that continues to be a driving force in modern neuroscience research.
A central tenet of this initiative has been high-resolution mapping of neurons and neuronal connectivity in the brain. The small size and structural detail of neurons has necessitated the use of electron microscopy for fine mapping approaches. This has been met with significant developments over the last few years in higher resolution and more sophisticated detection capabilities. Despite this, progress in high-throughput large-scale imaging processes has been limited, and true 3-dimensional brain mapping achievements have remained elusive.
Several technologies have sought to accelerate this initiative. Zeiss, for example, has pioneered the first commercial multi-beam scanning electron microscope – Zeiss MultiSEM. This technology was applied to the first primary challenge in the brain mapping initiative – characterization of a cubic millimeter of brain tissue, a volume believed to contain at least one cortical column and a basic computational module. From there scientists believe structural information can be built up and, along with functional data, can be used to map key features and ultimately neuronal connectivity.
To reliably image subcellular elements of neurons, a pixel resolution of 10 nm is required. The most established EM method for this scale has been block-face imaging with either focused ion beam ablation or in situ microtome cutting. Aside from the fact that these techniques are challenged by edge distortions and irreversible destruction of the samples, the logistics of integrating the 20,000 mm2 images for 3-D rendering are mind blowing – 50 ns per pixel or 6 years per cubic millimeter.
Multi-beam SEM theoretically provides an answer this problem, although, until the advent of the Zeiss MultiSEM, instrument developments remained largely in the prototype stage. The MultiSEM is similar in overall principle to single-beam SEM – the sample is bombarded with a focused beam probe about 1 nm in size. The electron beam is scanned line by line over the sample, ultimately generating the SEM image.
The MultiSEM 505 accelerates the process through parallelization. The beam produces 61 electron beams which in turn produce secondary electrons reflected from the image and projected onto a multi-detector, one beam per channel. This parallel data capture and processing can cut the logistics time interval to 3 or 4 months instead of 6 years with single SEM. The Zeiss MultiSEM 506 expands this to 91 beams and a field of view 3.5 times larger, further accelerating the imaging process.
At the Society for Neuroscience Zeiss press conference, topics encompassed not only the achievements of the MultiSEM, but the fact that large-scale data acquisition is now met with the next challenge in 3-D brain mapping – the big data issue. Each scan results in 1 petabyte of data. Considering the mouse brain is roughly 10x10x5 cubic mm and 80 million neurons in size, the total data set would reach 1.5 exabytes. An exabyte is 1,000 petabytes, and a petabyte is 1,000,000 bytes. Realistically, there is not much to compare exabytes with, in fact it is estimated that 5 exabytes is equal to all of the words spoken by mankind, and more that the world's current data storage capabilities. Not big but immense data…
An important next step in brain mapping and MultiSEM technology will be discerning useful data from background data such that only the most relevant details are processed. This would represent a huge leap in imaging efficiency and would greatly accelerate the BRAIN initiative. Not an easy task, this area will undoubtedly remain challenging and exciting for some time to come.
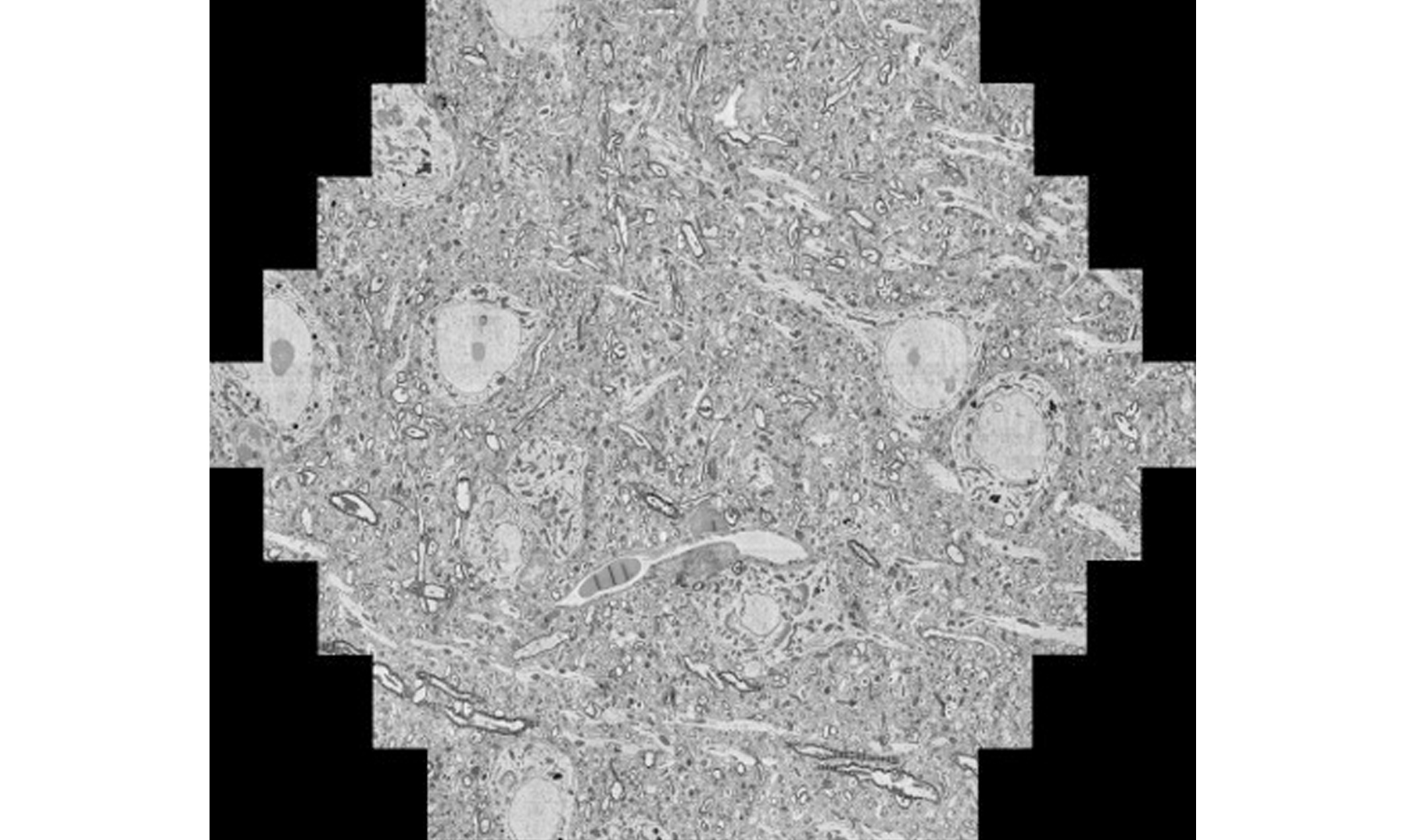
A reconstructed slice image of mouse brain taken from ZEISS multiSEM. Zeiss: https://stories.zeiss.com/en/a-20000-piece-puzzle-but-no-model/

A reconstructed 3-D mapping image of the neocortex. Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, et al. Saturated Reconstruction of a Volume of Neocortex. Cell 2015;162:648–61. doi:10.1016/j.cell.2015.06.054